CAPRI COVID-19 Round 51 - Target 183
This page provides information about the predictions and analyses of the CAPRI COVID Round 51, target T183.
Methodology
The results presented are of the Scorer submissions of Target 183.
Contacts: Contacts between entities were determined using a 5 Å distance threshold. Clashes (contacts below 2.5 Å), if any, were ignored. Only inter-chain contacts were considered.
Only heteromeric contacts were kept. Then, for all models, we counted the number of contacts (defined as “contact hits”), the number of contacts a residue makes at the interface (defined as “residue hits”), and the number of different residues a residue interacts with (defined as “distinct contacts”).
Sequence conservation: Sequence conservation was calculated using Rate4Site. Here, a low score means good conservation.
Target 183
The set consists of 164 models.
All the downloadable file can be found at the end of this page.
Target description
| Type | Ligand | Receptor | ||
|---|---|---|---|---|
| Species | SARS-CoV-2 | Homo sapiens (Human) | ||
| Name | Nsp8 | EXOSC8 | ||
| Uniprot ID | P0DTD1 | Q96B26 | ||
| PDB Template | 2NN6 | 3UB0.D, 2AHM.G, 6XIP | ||
| Description | Multifunctional protein involved in the transcription and replication of viral RNAs. Contains the proteinases responsible for the cleavages of the polyprotein. | Non-catalytic component of the RNA exosome complex which has 3’->5’ exoribonuclease activity and participates in a multitude of cellular RNA processing and degradation events. In the nucleus, the RNA exosome complex is involved in proper maturation of stable RNA species such as rRNA, snRNA and snoRNA, in the elimination of RNA processing by-products and non-coding ‘pervasive’ transcripts, such as antisense RNA species and promoter-upstream transcripts (PROMPTs), and of mRNAs with processing defects, thereby limiting or excluding their export to the cytoplasm. |
Analysis of viral protein
Visualization
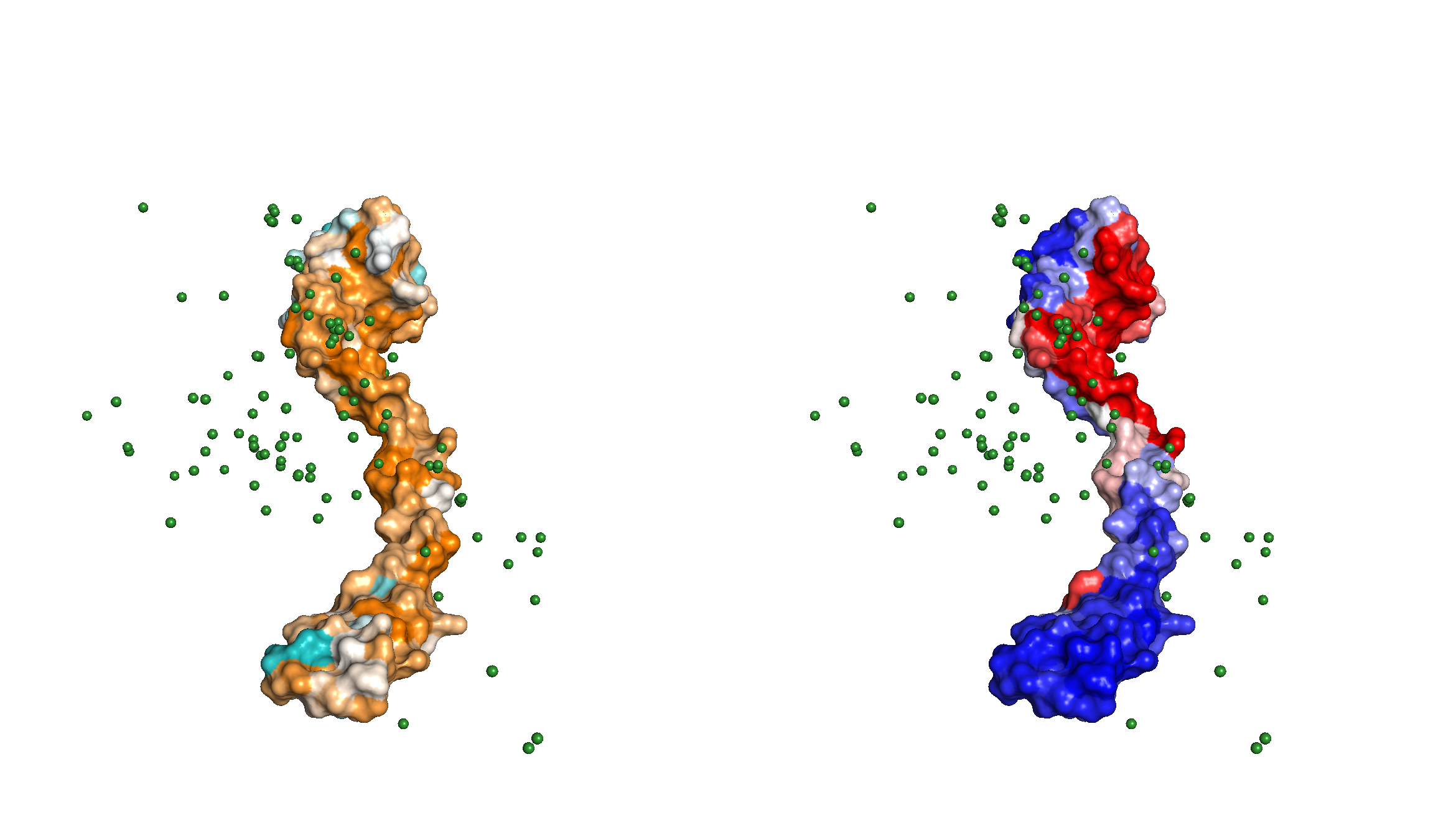
Surface representation of the ligand Nsp8 protein sequence conservation (left), coloring from orange (conserved) to teal (not conserved) and of the residue hits (right), colored from red (high occurrence, capped at the 90th percentile for better visualization) to blue (few occurrences). Green spheres are the center of mass of receptors for every model.
The PyMOL session where this image comes from can be downloaded here
Residue hit and conservation plot
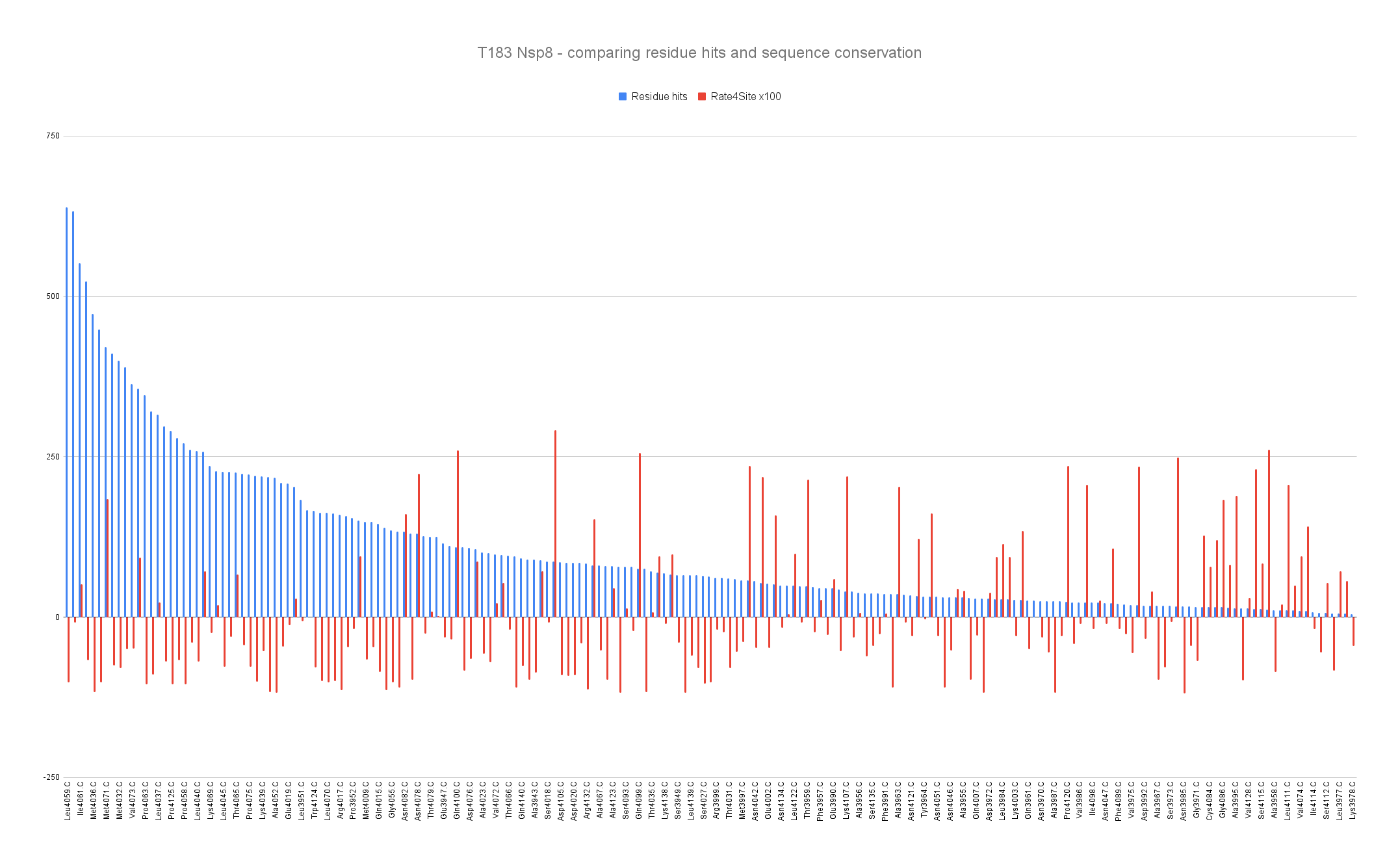
Barplot representing the residue hits (blue) and the sequence conservation (red) with Rate4Site score where the lowest means the most conserved of the Nsp8 residues.
Analysis of human protein
Visualization
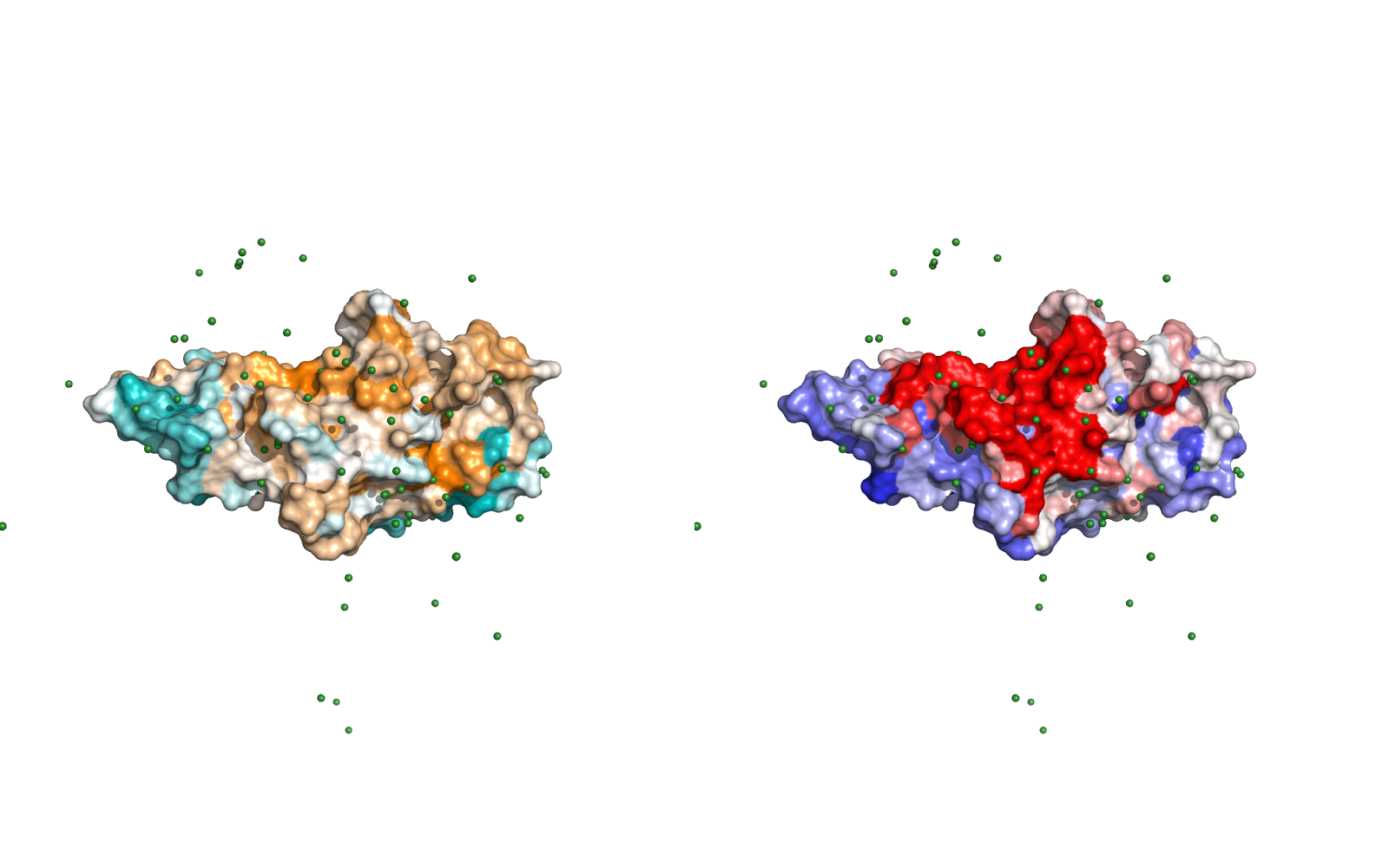
Surface representation of the ligand EXOSC8 protein sequence conservation (left), coloring from orange (conserved) to teal (not conserved) and of the residue hits (right), colored from red (high occurrence, capped at the 90th percentile for better visualization) to blue (few occurrences). Green spheres are the center of mass of receptors for every model.
The PyMOL session where this image comes from can be downloaded here
Residue hit and conservation plot
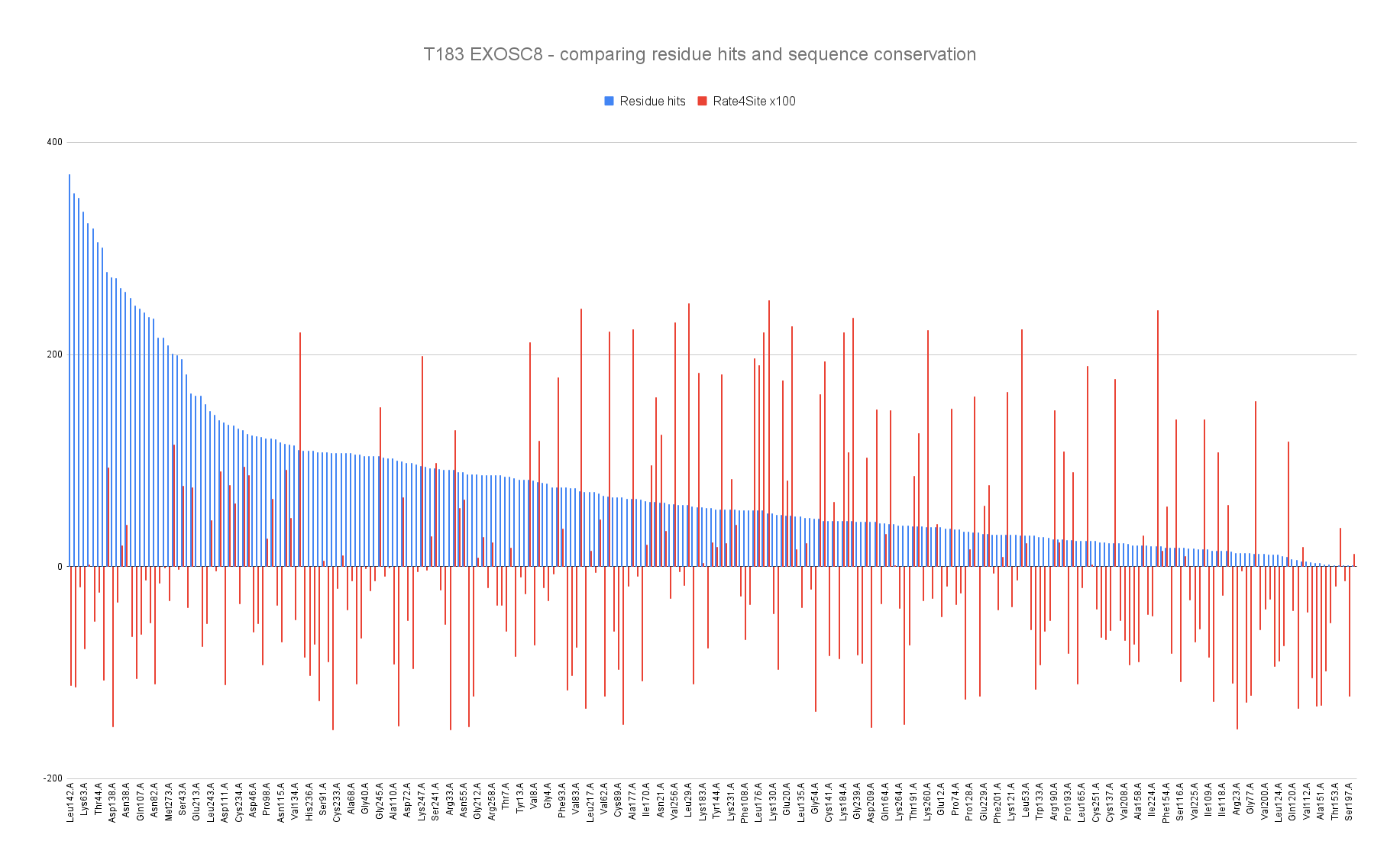
Barplot representing the residue hits (blue) and the sequence conservation (red) with Rate4Site score where the lowest means the most conserved of the EXOSC8 residues.
Downloadable files
| Target | PyMOL sessions | Contact hits | CSV files for receptor and ligand |
|---|---|---|---|
| T183 | T183_ligand / T183_receptor | T183_contact-hits | T183_csv |
The description of the dowloadable files can be found here
For questions, comments or feedback please contact Marc Lensink or Théo Mauri

