CAPRI-COVID-19 Round 51 - Expanded target information
CASP14 target H1103; CAPRI target T181
This is a complex of the SARS-COV2 protein Orf3a (PDB:6xdc) with the Human heme oxygenase HMOX1 (PDB:1n3u): A classical docking target, (A1B1 stoichiometry), with known structures of both components.
Sequence ORF3a
>C1905 ORF3a, SARS-CoV-2; PDB:6xdc
MDLFMRIFTIGTVTLKQGEIKDATPSDFVRATATIPIQASLPFGWLIVGVALLAVFQSASKIITLKKRWQ
LALSKGVHFVCNLLLLFVTVYSHLLLVAAGLEAPFLYLYALVYFLQSINFVRIIMRLWLCWKCRSKNPLL
YDANYFLCWHTNCYDYCIPYNSVTSSIVITSGDGTTSPISEHDYQIGGYTEKWESGVKDCVVLHSYFTSD
YYQLYSTQLSTDTGVEHVTFFIYNKIVDEPEEHVQIHTIDGSSGVVNPVMEPIYDEPTTTTSVPL
Sequence HMOX1
>sp|P09601|HMOX1_HUMAN Heme oxygenase 1 OS=Homo sapiens; PDB:1n3u
MERPQPDSMPQDLSEALKEATKEVHTQAENAEFMRNFQKGQVTRDGFKLVMASLYHIYVALEEEIERNKE
SPVFAPVYFPEELHRKAALEQDLAFWYGPRWQEVIPYTPAMQRYVKRLHEVGRTEPELLVAHAYTRYLGD
LSGGQVLKKIAQKALDLPSSGEGLAFFTFPNIASATKFKQLYRSRMNSLEMTPAVRQRVIEEAKTAFLLN
IQLFEELQELLTHDTKDQSPSRAPGLRQRASNKVQDSAPVETPRGKPPLNTRSQAPLLRWVLTLSFLVAT
VAVGLYAM
CAPRI Target T182
A: Nsp15 - B: NUTF2, Probable stoichiometry: A1B2
NUTF2 /NFT2 is a small (127 aa) hub protein (43 known interactions in IntAct), related to nuclear import.
The two known functions of Nsp15 (endoribonuclease, and interfering with dsRNA-interacting proteins) do not seem to directly correlate with the function of NUTF2.
There is no obvious reason to assume an obligate participation of a third molecule.
Roland Dunbrack’s analysis indicates that NUTF2/NTF2 is a dimer.
- Interface residues of the NUTF2 homodimer, should be indicated.
- Previous studies have identified a surface hydrophobic patch centred on Trp7 in rNTF2 (rat protein) (Phe5 in yNTF2: Yeast protein) that interacts with FxFG repeats.
- According to the rat structure, the FxFG peptide binds to the subunit interface residues: Pro75, Pro77 and TRP7 contribute to the hydrophobic depression, whereas in the second chain Glu35, T115, Cys38, Gln45 and Gln47 contribute. Not totally sure about alignment with Rat sequence (had to derive the alignment manually).
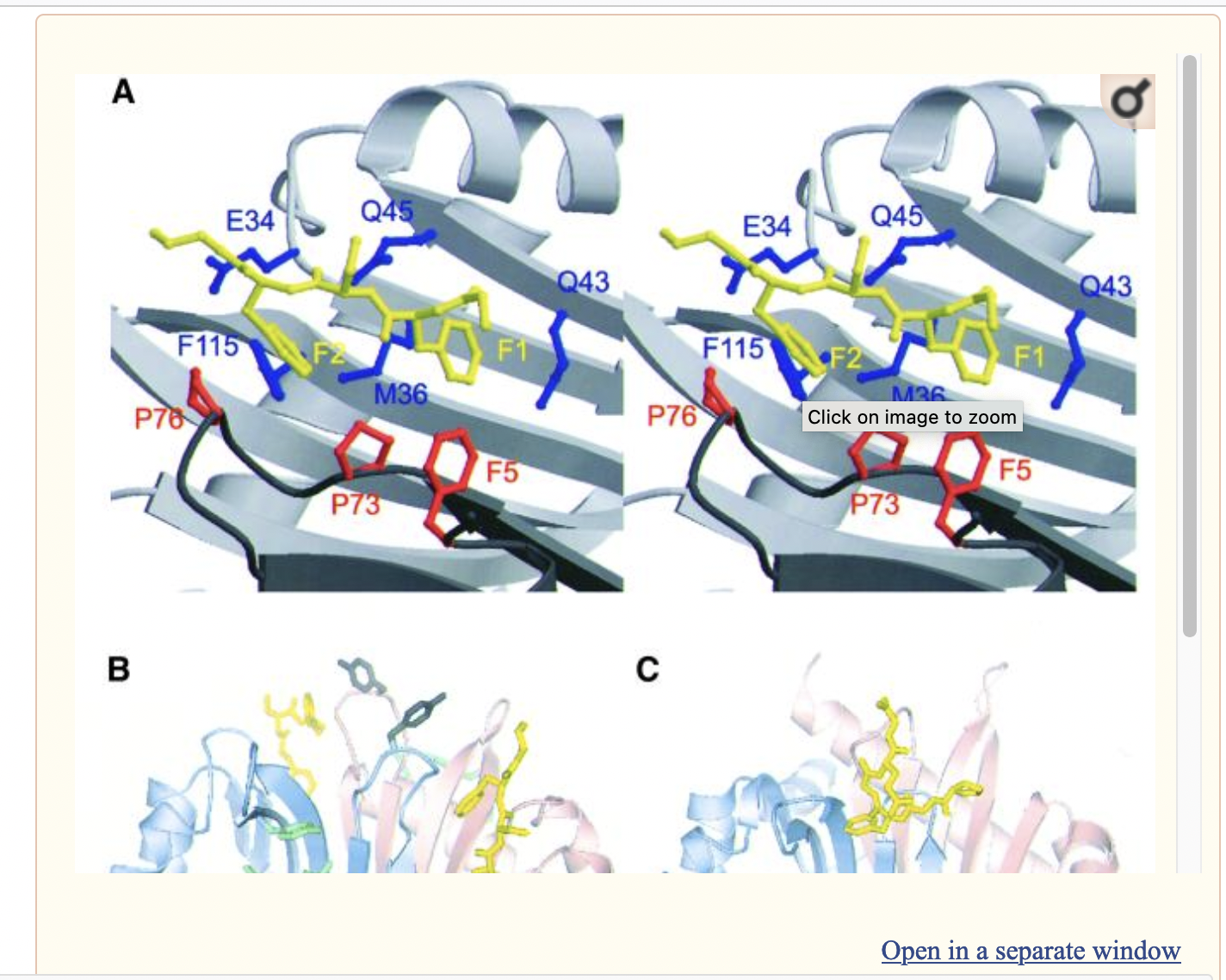
Figure 2 from EMBO J. 2002 Jun 17; 21(12): 2843–2853. doi: 10.1093/emboj/cdf305
Active site residues of NSP15 the endoribonuclease are: His235, His250, Lys290, Thr341, Tyr343, and Ser294. Structure of SARS-COV-2 NSP15, solved 2020 (6VWW) and then by cryo-EM (2021) (see f.e. 7K0R) , the protein is a homo-hexamer (D3 symmetry). Interfaces with other subunits of NSP15 should not be accessible for interaction with the NUTf2 dimer.
Sequence SARS-COV-2 Nsp15
>sp|P0DTD1|6453-6798
SLENVAFNVVNKGHFDGQQGEVPVSIINNTVYTKVDGVDVELFENKTTLPVNVAFELWAK
RNIKPVPEVKILNNLGVDIAANTVIWDYKRDAPAHISTIGVCSMTDIAKKPTETICAPLT
VFFDGRVDGQVDLFRNARNGVLITEGSVKGLQPSVGPKQASLNGVTLIGEAVKTQFNYYK
KVDGVVQQLPETYFTQSRNLQEFKPRSQMEIDFLELAMDEFIERYKLEGYAFEHIVYGDF
SHSQLGGLHLLIGLAKRFKESPFELEDFIPMDSTVKNYFITDAQTGSSKCVCSVIDLLLD
DFVEIIKSQDLSVVSKVVKVTIDYTEISFMLWCKDGHVETFYPKLQ
Sequence NUFT2/NTF2
>sp|P61970|NTF2_HUMAN Nuclear transport factor 2 OS=Homo sapiens OX=9606 GN=NUTF2 PE=1 SV=1
MGDKPIWEQIGSSFIQHYYQLFDNDRTQLGAIYIDASCLTWEGQQFQGKAAIVEKLSSLP
FQKIQHSITAQDHQPTPDSCIISMVVGQLKADEDPIMGFHQMFLLKNINDAWVCTNDMFR
LALHNFG
CAPRI target T183
A: Nsp8 – B: EXOSC8; probable stoichiometry A1B1
EXOSC8 is a non-catalytic component of the RNA exosome complex (9 subunits).
PDB:2NN6 seems to contain the whole exosome. Q13868 (EXOSC2) and Q9NPD3 (EXOSC4) and they both show up in UCSF High confidence dataset, with Nsp8 (Gordon et al., 2020). Both proteins do not show up using Nsp7 or Nsp12. as baits in this study.
On the other hand: Nsp8-EXOSC2, Nsp8-EXOSC3 and Nsp8-EXOSC5 are in fact part of the 80 % coverage list (on shared Google Drive).
Note that interactions with these other subunits of the complex could be ‘carry overs’ e.g.: NSP8 could physically bind to any one of these subunits of the Exosome, whereas other subunits could show up as ‘binders’ only because they are part of the full complex.
EXOSC8: Residues to be occluded for docking on this subunit, should be all the residues that are part of the interfaces of this subunit with other members of the complex (e.g. representing multiple interfaces). A full analysis of the binding interfaces of NSP8 needs to be done to enumerate these residues.
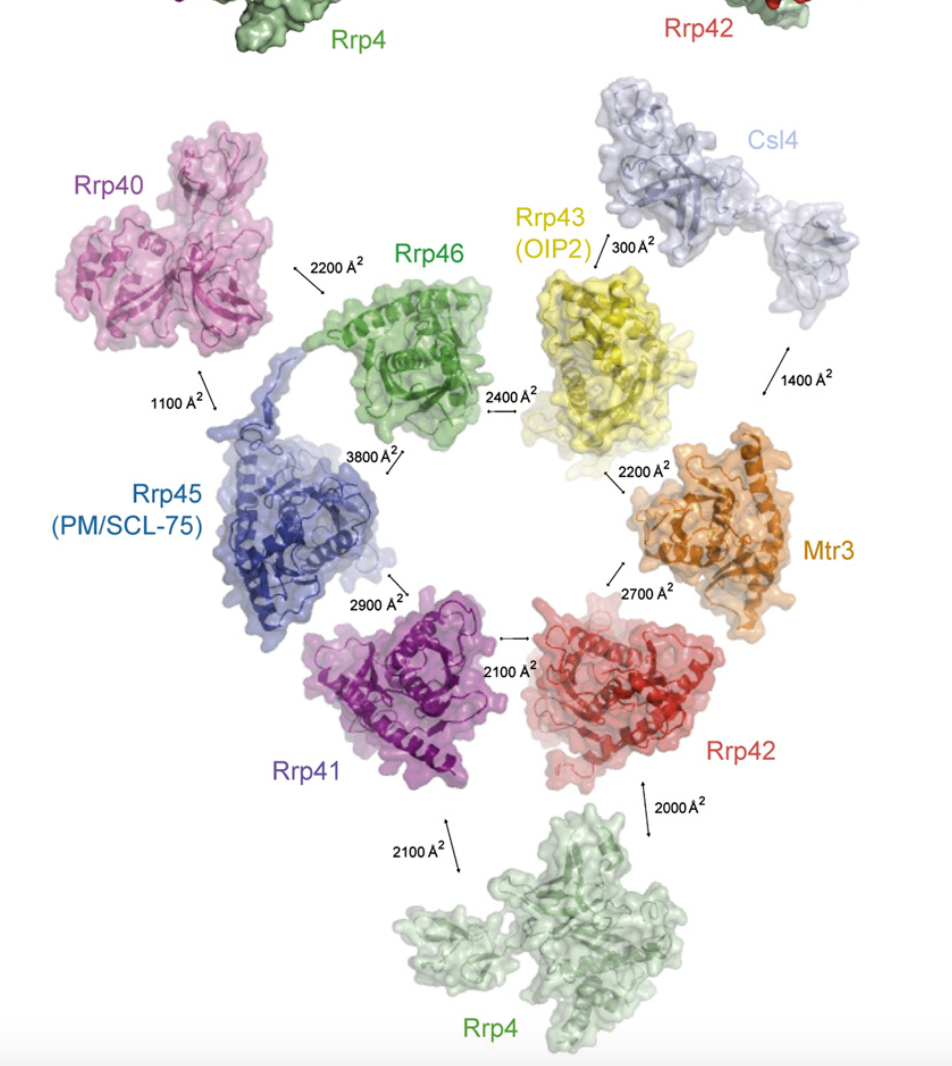
The Figure below (copied from:https://doi.org/10.1016/j.cell.2006.10.037) shows the buried areas (interfaces) between the 9 subunits in the reconstituted Exosome.
These contacts/ interfaces may (eventually) be used to delineate the available regions of ExosC8 (Rrp43), available for interaction with NSP8. FYI: EXOSC2= Rrp4,EXOSC3=Rrp40, EXOSC4=Rrp41, EXOSC5= Rrp46.
Sequence SARS-COV-2 NSP8 (198 residues)
>sp|P0DTC1|3943-4140
AIASEFSSLPSYAAFATAQEAYEQAVANGDSEVVLKKLKKSLNVAKSEFDRDAAMQRKLE
KMADQAMTQMYKQARSEDKRAKVTSAMQTMLFTMLRKLDNDALNNIINNARDGCVPLNII
PLTTAAKLMVVIPDYNTYKNTCDGTTFTYASALWEIQQVVDADSKIVQLSEISMDNSPNL
AWPLIVTALRANSAVKLQ
Sequence Human EXOSC8 (Rrp43)
>sp|Q96B26|EXOS8_HUMAN Exosome complex component RRP43 OS=Homo sapiens OX=9606 GN=EXOSC8 PE=1 SV=1
MAAGFKTVEPLEYYRRFLKENCRPDGRELGEFRTTTVNIGSISTADGSALVKLGNTTVIC
GVKAEFAAPSTDAPDKGYVVPNVDLPPLCSSRFRSGPPGEEAQVASQFIADVIENSQIIQ
KEDLCISPGKLVWVLYCDLICLDYDGNILDACTFALLAALKNVQLPEVTINEETALAEVN
LKKKSYLNIRTHPVATSFAVFDDTLLIVDPTGEEEHLATGTLTIVMDEEGKLCCLHKPGG
SGLTGAKLQDCMSRAVTRHKEVKKLMDEVIKSMKPK
CAPRI target T184
A: Nsp7- B: RhoA ; probable stoichiometry: A1 B1
RhoA is a small GTPase that has GTP-bound and GDP-bound forms, with many post-translational modifications. Signalling/hub protein (27 curated interactions, plus it can be a subunit of a dozen more complexes), involved in cytoskeleton reorganization. Looks to be always a monomer in its protein-protein interactions. It has known interactions with viral proteins. A literature search might shed more light on the exact RhoA form and if there are additional proteins/molecules involved. To be seen if the interaction is just with Nsp7 or also with Nsp8/Nsp12.
Other relate small GTPases: Rab10/Rab1A/Rab18/Rab8A/Arf6/Rab7A/RalA
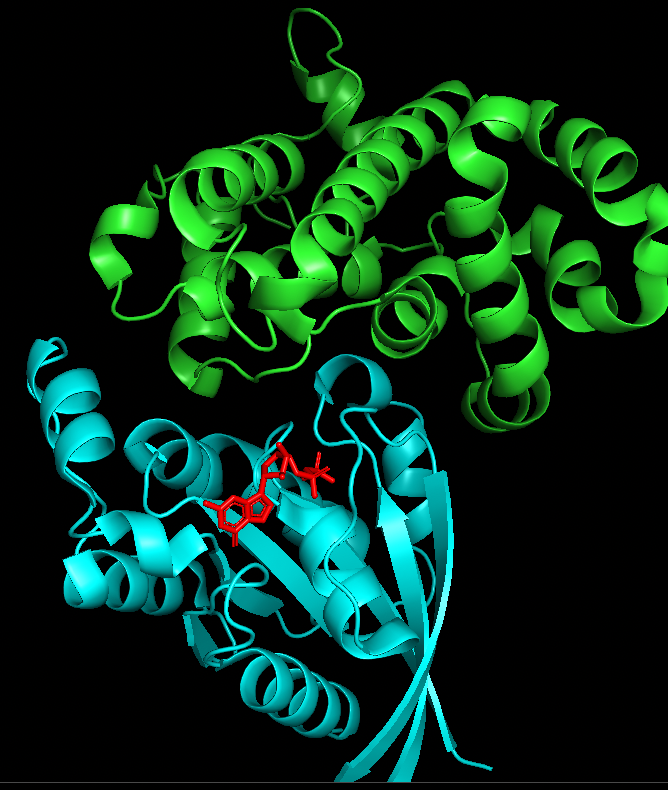
The above Picture is a screenshot from Pymol of 1OW3 (RhoA.GDP.MgF3-in Complex with RhoGAP; both Human). RhoA interacts with many different proteins forming large interaction interfaces. Some of these interfaces may engage the same surface region of RhoA. This needs to be examined to find out if this region (or similar) is also part of the interface with NSP7 in the submitted models.
Sequence SARS-COV-2 NSP7 (83 residues)
>sp|P0DTC1|3860-3942
SKMSDVKCTSVVLLSVLQQLRVESSSKLWAQCVQLHNDILLAKDTTEAFEKMVSLLSVLL
SMQGAVDINKLCEEMLDNRATLQ
Sequence human RhoA
>sp|P61586|RHOA_HUMAN Transforming protein RhoA OS=Homo sapiens OX=9606 GN=RHOA PE=1 SV=1
MAAIRKKLVIVGDGACGKTCLLIVFSKDQFPEVYVPTVFENYVADIEVDGKQVELALWDT
AGQEDYDRLRPLSYPDTDVILMCFSIDSPDSLENIPEKWTPEVKHFCPNVPIILVGNKKD
LRNDEHTRRELAKMKQEPVKPEEGRDMANRIGAFGYMECSAKTKDGVREVFEMATRAALQ
ARRGKKKSGCLVL
For questions, comments or feedback please contact Marc Lensink or Théo Mauri

